- Technique of using live organisms or enzymes from organisms to produce products and processes useful to humans.
- eg. Invitro fertilizations, synthesizing gene and using it, developing DNA vaccine.
- According to European Federation of Biotechnology (EFB), biotechnology is - ‘The integration of natural science and organisms, cells and molecular analogues for products and services.’
Principles of Biotechnology:
(i) Genetic engineering: Alter chemistry of genetic material (DNA and RNA) to introduce them into host organisms and change host’s phenotype.
(ii) Bioprocess engineering: Maintenance of sterile condition for growth of desired cell in large quantities.
Development of principles of genetic engineering:
- Traditional hybridization often led to inclusion of undesired genes along with desired genes, whereas genetic engineering allows to isolate desirable genes without introducing undesirable genes.
First construction of artificial recombinant DNA: By Stanley Cohen and Hebert Boyer.
- In 1972, they isolated antibiotic resistance gene by cutting out DNA piece from plasmid (autonomously replicating circular extra chromosomal DNA) of Salmonella typhimurium.
- The cut piece is then linked to plasmid DNA which act as vector and transfers alien piece of DNA to host. Now, it replicates using new host’s DNA polymerase and make multiple copies.
- This combination of circular autonomously replicating DNA created invitro by linking cut DNA with plasmid -recombinant DNA.
Tools of Recombinant DNA Technology:
1. Restriction enzymes: Cuts DNA at specific locations (molecular scissors).
- In 1963, 2 enzymes for restricting growth of bacteriophage in Escherichia coli were isolated. 1 of them added methyl group to DNA and other cut DNA
- Hind II: First restriction endonuclease which cut DNA by recognising specific sequence of 6 base pairs (recognition sequence)
- There are more than 900 restriction enzymes isolated from over 230 strains.
- In Eco RI from Escherichia coli RY 13, ‘E’ comes from genus (Escherichia), ‘co’ from species (coli), ‘R’ is strain while ‘I’ is roman numeral in order in which enzyme were isolated from that strain of bacteria.
- They belong to larger class - nucleases
® Exonuclease: Remove nucleotides from ends of DNA.
® Endonuclease: Cuts at specific positions within DNA.
- It functions by inspecting DNA length, recognises specific palindromic nucleotide sequence in DNA (sequence that reads same on 2 strands of DNA when orientation of reading is kept same)
- eg. 5' ® GAATTC ® 3' 3' ® CTTAAG ® 5'
- It cuts DNA little away from centre but between 2 same bases on opposite strands, which leave single stranded portions. Each strand has overhanging stretches - sticky ends (form H bonds with complementary cut part).
- These sticky ends are joined by DNA ligase, forming recombinant DNA molecule.
- Unless vector and source DNA aren’t cut by same restriction enzyme, recombinant molecule can’t be created.
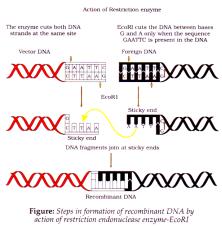
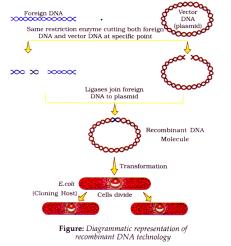
Separation and Isolation of DNA fragments:
- Cutting of DNA by restriction endonuclease results in fragments of DNA which can be separated by gel electrophoresis.
- DNA are negatively charged molecule, so move to anode under electric field, through matrix agarose (natural polymer extracted from sea weeds).
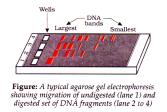
- Fragments get separated according to size. Smaller the fragment, farther it moves.
- These are visualised by staining in ethidium bromide followed by UV exposure. Bright orange bands are visible.
- The separated bands are cut out from gel and extracted called elution.
2. Cloning Vectors: For many copies, vector should have high copy number.
- Help in easy linking of foreign DNA and selection of recombinants from non-recombinants.
- Bacteriophage (high no./ cell) have high copy numbers whereas some plasmids have 1 - 2 copies/ cell and some have 15 - 100 copies/cell. They both have ability to replicate within bacterial cell independent of control of chromosomal DNA.
- Features facilitating cloning into vector.
- Origin of replication: Sequence where replication starts.
- Controls copy no. of linked DNA so if many copies of target DNA are required, it should be cloned in vector whose origin support high copy number.
3. Selectable marker: Help in identifying and eliminating non transformants and selectively permitting growth of transformants.
eg. genes encoding resistance to antibiotics - Ampicillin, chloramphenicol, tetracycline or kanamycin for
E. coli (Normal E. coli doesn’t carry resistance to any).
Transformation - Process by which DNA piece is introduced in host.
4. Cloning sites:
- Ligation of foreign DNA needs single recognition site present in 1 out of 2 antibiotic resistance genes.
- eg. legating foreign DNA at Bam H I site of tetracycline resistance gene in vector pBR 322 results in loss of tetracycline resistance in recombinant plasmid, but still can be selected from non-recombinant by placing transformants on tetracycline medium.
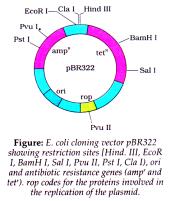
- Recombinants will grow on ampicillin medium but not on tetracycline medium. Non-recombinants grow on medium containing both antibiotics.
- One antibiotic resistance gene helps in selecting transformants whereas other gets inactivated due to alien DNA.
Drawback: Cumbersome process as it requires simultane-ous placing on 2 plates having different antibiotics.
Another selectable marker: Differentiate recombinants from non-recombinants on basis of ability to produce colour in presence of chromogenic substrate .
- r. DNA is inserted within coding sequence of b galactosidase which results in inactivation of gene for enzyme synthesis - insertional inactivation.
- If plasmid doesn’t have insert, chromogenic substrate gives blue colour colonies.
- Presence of insert results in insertional inactivation and colonies don’t produce any colour, these are transformants I recombinants.
5. Vectors for cloning genes in plants and animals:
- Agrobacterium tumifaciens: Its a pathogen of dicot plants that delivers DNA piece - T - DNA to transform normal plant cells to tumor and direct them to produce chemicals required by pathogen
- Retrovirus: In animals they have ability to transform normal cells to cancerous cells.
- But, now these are modified into cloning vectors which is not more pathogenic or harmful and helps to deliver desirable genes into host.
3. Competent Host: For transformation with r. DNA.
- DNA is hydrophillic, so can’t pass through membrane.
- To make bacterial cell competent to take up DNA, we treat it with specific concentration of divalent cation (Ca+2) which increases efficiency with which DNA enters bacterium through pores in wall.
- rDNA is then forced into such cells by incubating cells with rDNA on ice, followed by heat shock (42°C) and then putting them back on ice. This enables bacteria to take rDNA.
Other methods:
- Micro-injection: r. DNA is directly injected into nucleus of animal cell.
- Biolistics/gene gun: Cells are bombarded with high velocity micro particles of gold or tungsten coated with DNA, suitable for plants.
- Disarmed pathogen: Which when allowed to infect cell transfers rDNA into host.
Process of recombinant DNA technology:
1. Isolation of the Genetic Material (DNA):
- Nucleic acid is genetic material without exception.
- In order to cut DNA, it needs to be in pure form, free from macro-molecules, cell has to break open to release DNA along with RNA, proteins, polysaccharides and lipids.
- This is done by treating cells with enzymes -lysozyme (bacteria), cellulase (plant cells), chitinase (fungus) and purified DNA precipitates out after adding chilled ethanol (as fine threads in suspension).
- RNA is removed by ribonuclease, protein by protease etc.
2. Cutting of DNA at specific locations:
- Restriction endonuclease cut source DNA and vector DNA at specific position at optimal conditions. Gel electrophoresis is employed to check progression of restriction enzyme digestion.
- Cut out gene of interest from source and vector DNA is mixed and ligase is added which results in recombinant DNA.
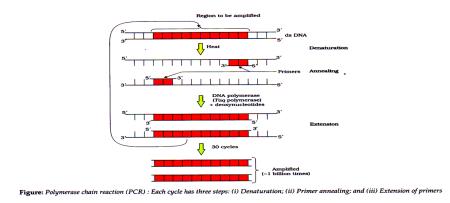
3. Amplification of Gene of Interest using PCR: Polymerase Chain Reaction.
- Multiple copies of ‘gene of interest’ is synthesised in vitro using 2 sets of primers (small chemically synthesised oligonucleotides complementary to DNA region) and enzyme DNA polymerase.
- Enzyme extends primers using nucleotides in reaction and genomic DNA as template. If replication of DNA is repeated, a billion copies of DNA segment can be made, by the use of thermostable DNA polymerase (isolated from bacterium, Thermus aquaticus) which is active at high temperature and denatures ds DNA.
- This fragment can be ligated with vector for further cloning.
4. Insertion of Recombinant DNA into Host cell/organism:
- Recipient cell should be made competent to receive DNA.
- If r. DNA bearing gene for ampicillin resistance is transferred to E. coli, host cell become resistant to ampicillin.
- If transformed cells are spread on agar plates having ampicillin, only transformants grow. So, due to ampicillin resistance gene, one is able to select transformed cell hence this gene is selectable marker.
5. Obtaining the foreign gene product:
- Recombinant protein: Protein encoding gene expressed in heterologous host.
- Cells harbouring cloned genes of interest may be grown on small scale in laboratory.
- They can also be multiplied in continuous culture system where used medium is drained from one side while fresh one is added from other side to maintain cells in active log/ exponential phase. It yields higher amount of desired protein.
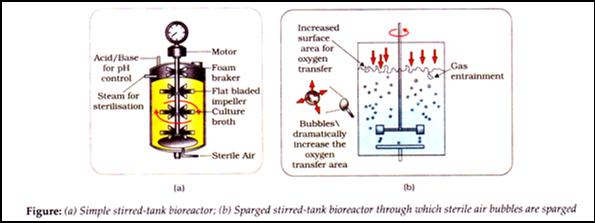
- Bioreactors: Vessels in which raw material are biologically converted into specific products using microbial plant, human cells etc. and large volumes (100 - 1000 Iitres) of culture can be processed by providing optimal conditions (pH, temperature, substrate, salts, vitamins, oxygen). Most common is stirring type.
- Stirred tank: Usually cylindrical/ curved base to facilitate even mixing of contents, facilitates oxygen availability (air can be bubbled through reactor), it has agitator system, oxygen delivery system, foam control system, temperature and pH control system and sampling port to withdraw small volumes periodically.
6. Downstream Processing:
- Product is subjected to processes like separation and purification.
- It has to be preserved in suitable solution and undergoes clinical trials (in case of drugs).
- Quality control testing is also required.


… [Trackback]
[…] Read More Info here on that Topic: eklabhyaclasses.com/blog/biotechnology-and-principle/ […]
… [Trackback]
[…] Here you can find 91532 additional Information to that Topic: eklabhyaclasses.com/blog/biotechnology-and-principle/ […]
… [Trackback]
[…] Find More on that Topic: eklabhyaclasses.com/blog/biotechnology-and-principle/ […]
… [Trackback]
[…] Find More here to that Topic: eklabhyaclasses.com/blog/biotechnology-and-principle/ […]
… [Trackback]
[…] Read More on that Topic: eklabhyaclasses.com/blog/biotechnology-and-principle/ […]
… [Trackback]
[…] Information on that Topic: eklabhyaclasses.com/blog/biotechnology-and-principle/ […]